



whole genome, one cell at a time
Explore Our Research
Embryos start out life as a single cell but quickly diversify into the bones, muscles, neurons, and skin that make up a plant or animal. Our lab is interested in how these diverse cell types arise. Generally, we address these and other developmental biology questions using cutting-edge technologies like single-cell-resolution genomics and single-molecule imaging. Our research combines hypothesis-driven molecular experimentation and computational analysis.
We use the microscopic nematode worm, Caenorhabditis elegans, as a genetic model. C. elegans are popular among researchers for their rapid and invariant embryonic development that can be viewed under the microscope over the course of a day. In addition, C. elegans were the first animal species to have their genome fully sequenced. Selected for their small genome size in that project, they remain a fantastic model of genome-wide gene expression and regulation.
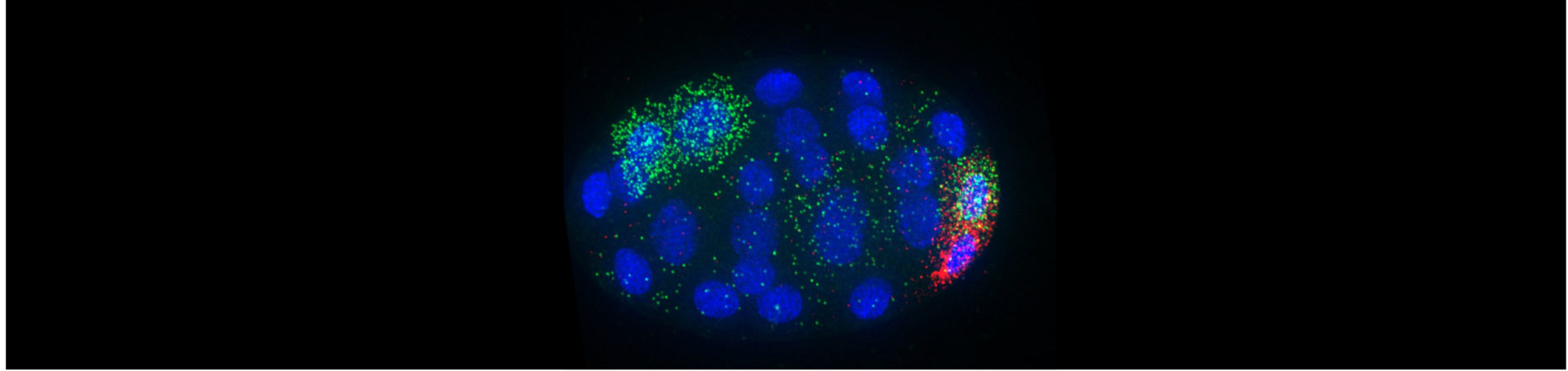
C. elegans mRNA's localize subcellularly in early embryos
The field has the ultimate goal of characterizing gene expression dynamics in developing C. elegans embryos in an effort to understand how specific cohorts of genes drive cellular differentiation. Towards this effort, Dr. Nishimura characterized the individual transcriptomes of the first two zygotic daughter cells using single-cell RNA-seq [Osborne Nishimura et al., 2015]. She later contributed to a collaborative effort to characterize the transcriptomes of all cells produced through the first four cell divisions [Tintori et al., 2016]. Interestingly, the transcriptomes of even the earliest C. elegans cells begin to diversify despite the fact that de novo transcription does not initiate at those stages.
Prior to the onset of transcription, how do differences in mRNA abundance arise? Our group hypothesizes that post-transcriptional mechanisms such as differential mRNA stabilization, transport, inheritance, or decay drive the patterning we have found. Our research seeks to determine the mechanisms driving these cell-specific mRNA patterns and to understand how differentially abundant transcripts lead to lineage-specific differentiation.erm-1_projection
We have discovered that mRNA transcripts with cell-specific patterns of enrichment often accumulate in interesting regions within the cell! This was a big surprise.
We recently discovered that mRNA transcripts that undergo no or low translation are sequestered into biomolecular condensates (P granules) as a second step following their down-regulation. Current work in the lab is investigating mechanisms that direct mRNA localization to membranes and the links between those membrane localizations and regulatory control.
C. elegans intestinal development - a model of organogenesis
All intestines digest food, but the intestine of the C. elegans nematode worm undertakes additional tasks that, in humans, are performed by separate organs. This includes coordinating fat storage, growth, genetic aging, environmental stress, and response to infection. At only 20-cells, the C. elegans intestine is a model of developmental genetics. In it, a network of GATA transcription factors directs intestinal cell fate by culminating in the expression of ELT-2 (Erythroid Like Transcription factor), the major intestinal regulator, and its partner ELT-7. This network has been instrumental in revealing general aspects of network biology, but critical gaps remain and we have yet to satisfyingly link it to the various tasks the intestine accomplishes once formed. Our research seeks to rectify unexplained aspects of the regulatory network and to expand the network to integrate stages of development when the intestine performs multiple functions.
High-resolution transcriptomics in cancer and neurobiology
WHAT WE ASK
Embryos start out life as a single cell but quickly diversify into cells of many types such as those that comprise our muscles, neurons, and skin. How do these diverse cell types arise? Our lab aims to better understand how cohorts of genes orchestrate these developmental processes. In the process, we discover novel modes of genetic control.
HOW WE ASK IT
We address biological questions using cutting-edge technologies like single-cell-resolution genomics and single-molecule imaging. Our research combines hypothesis-driven molecular experimentation and computational analysis. Researchers in our group are trained in genetics, molecular biology, and computational biology.
MEET THE WORMS
The cellular machinery used to regulate gene expression is well conserved across animals. For this reason, we can capitalize on a powerful model system, the nematode worm Caenorhabditis elegans. These worms undergo rapid development in transparent eggs that can develop outside the mother. Each cell division occurs reproducibly and can be observed with detail. Further, we can disrupt each gene to study its precise function thanks to the efforts of a large community of C. elegans researchers who have worked together over many decades to ease discovery.

WHAT WE ASK
Embryos start out life as a single cell but quickly diversify into the bones, muscles, neurons, and skin that make up a plant or animal. How do these diverse cell types arise? Our lab is interested in how cohorts of genes orchestrate these developmental processes. In the process, we discover novel modes of genetic regulatory control.
HOW WE ASK IT
We address biological questions using cutting-edge technologies like single-cell-resolution genomics and single-molecule imaging. Our research combines hypothesis-driven molecular experimentation and computational analysis. Researchers in our group are trained in genetics, molecular biology, and computational biology.
MEET THE WORMS
We use the microscopic nematode worm, Caenorhabditis elegans, as a genetic model. C. elegans are popular among researchers for their rapid and invariant embryonic development that can be viewed under the microscope over the course of a day. C. elegans were the first animal species to have their genome fully sequenced and are excellent for use in genome-wide studies. Furthermore, a large community of C. elegans researchers has worked together to develop resources that allow for precise genetic manipulation and experimentation. Because the characteristics and machinery of gene regulatory control are shared across animals, the things we learn in this worm model are relevant to human health.
Research Projects
C. elegans mRNA's localize subcellularly in early embryos
The field has the ultimate goal of characterizing gene expression dynamics in developing C. elegans embryos in an effort to understand how specific cohorts of genes drive cellular differentiation. Towards this effort, Dr. Nishimura characterized the individual transcriptomes of the first two zygotic daughter cells using single-cell RNA-seq [Osborne Nishimura et al., 2015]. She later contributed to a collaborative effort to characterize the transcriptomes of all cells produced through the first four cell divisions [Tintori et al., 2016]. Interestingly, the transcriptomes of even the earliest C. elegans cells begin to diversify despite the fact that de novo transcription does not initiate at those stages.
Prior to the onset of transcription, how do differences in mRNA abundance arise? Our group hypothesizes that post-transcriptional mechanisms such as differential mRNA stabilization, transport, inheritance, or decay drive the patterning we have found. Our research seeks to determine the mechanisms driving these cell-specific mRNA patterns and to understand how differentially abundant transcripts lead to lineage-specific differentiation.
We have discovered that mRNA transcripts with cell-specific patterns of enrichment often accumulate in interesting regions within the cell! This was a big surprise.
We recently discovered that mRNA transcripts that undergo no or low translation are sequestered into biomolecular condensates (P granules) as a second step following their down-regulation. Current work in the lab is investigating mechanisms that direct mRNA localization to membranes and the links between those membrane localizations and regulatory control.
The field has the ultimate goal of characterizing gene expression dynamics in developing C. elegans embryos in an effort to understand how specific cohorts of genes drive cellular differentiation. Towards this effort, Dr. Nishimura characterized the individual transcriptomes of the first two zygotic daughter cells using single-cell RNA-seq [Osborne Nishimura et al., 2015]. She later contributed to a collaborative effort to characterize the transcriptomes of all cells produced through the first four cell divisions [Tintori et al., 2016]. Interestingly, the transcriptomes of even the earliest C. elegans cells begin to diversify despite the fact that de novo transcription does not initiate at those stages.
Prior to the onset of transcription, how do differences in mRNA abundance arise? Our group hypothesizes that post-transcriptional mechanisms such as differential mRNA stabilization, transport, inheritance, or decay drive the patterning we have found. Our research seeks to determine the mechanisms driving these cell-specific mRNA patterns and to understand how differentially abundant transcripts lead to lineage-specific differentiation.
We have discovered that mRNA transcripts with cell-specific patterns of enrichment often accumulate in interesting regions within the cell! This was a big surprise.
We recently discovered that mRNA transcripts that undergo no or low translation are sequestered into biomolecular condensates (P granules) as a second step following their down-regulation. Current work in the lab is investigating mechanisms that direct mRNA localization to membranes and the links between those membrane localizations and regulatory control.
All intestines digest food, but the intestine of the C. elegans nematode worm undertakes additional tasks that, in humans, are performed by separate organs. This includes coordinating fat storage, growth, genetic aging, environmental stress, and response to infection. At only 20-cells, the C. elegans intestine is a model of developmental genetics. In it, a network of GATA transcription factors directs intestinal cell fate by culminating in the expression of ELT-2 (Erythroid Like Transcription factor), the major intestinal regulator, and its partner ELT-7. This network has been instrumental in revealing general aspects of network biology, but critical gaps remain and we have yet to satisfyingly link it to the various tasks the intestine accomplishes once formed. Our research seeks to rectify unexplained aspects of the regulatory network and to expand the network to integrate stages of development when the intestine performs multiple functions.
Meet The Scientists
Lauren Billow
Undergraduate Researcher
Meet the scientists
Romario Romain
Technician
Camryn Daidone
Undergraduate Researcher
Meet the scientists
Meghan Costello, PhD
Postdoctoral Researcher
Annemarie Parker
Undergraduate Researcher
Lab Group Photos

Located at Colorado State University
in beautiful Colorado!